T1SEstacker
An Integrated Prediction Pipeline for Bacterial Type I Secreted Effectors.
Standalone Version of T1SEstacker:
User Guide
1. Introduction
This manual was prepared for the standalone version of T1SEstacker (version 1.0) and the related modules. T1SEstacker could be run as a whole pipeline. Alternatively, the modules could be used independently.
2. Manual of T1SEstacker
1) System requirement: Linux , Mac or windows.
2) Software or package prerequisites: The following software or packages should be pre-installed and configured into environmental variable path.
- Perl 5
- R==version 3.3 or later
- R package: randomForest, e1071
- *GO (version 1, if source code compilation needed)
- Python3 version==3.6-3.8
- Numpy>=1.19.4
- biopython>=1.78
- Pandas>=0.25.1
- Tensorflow>=2.4.0
3) Download and installation of T1SEstacker:
The T1SEstacker package (T1SEstacker.v1.0) could be downloaded in this page.
Decompress the “T1SEstacker.tar.gz” and get into ~/T1SEstacker/ from terminal. If the pre-compiled version does not work, try to re-compile all the GO scripts in the “codes” ,and replace the binary files in the “bin” sub-folder of each module with the newly compiled ones, respectively. Compiling GO scripts:
$ cd ~/T1SEstacker/MODULE_NAME/bin
$ go build ../codes/xxx.go
4) Input files:
There is a necessary protein sequence file .The ‘protein sequence file’ T1SEstacker requires them to be FASTA-formatted, as exemplified by the demonstrated “test.fasta”.
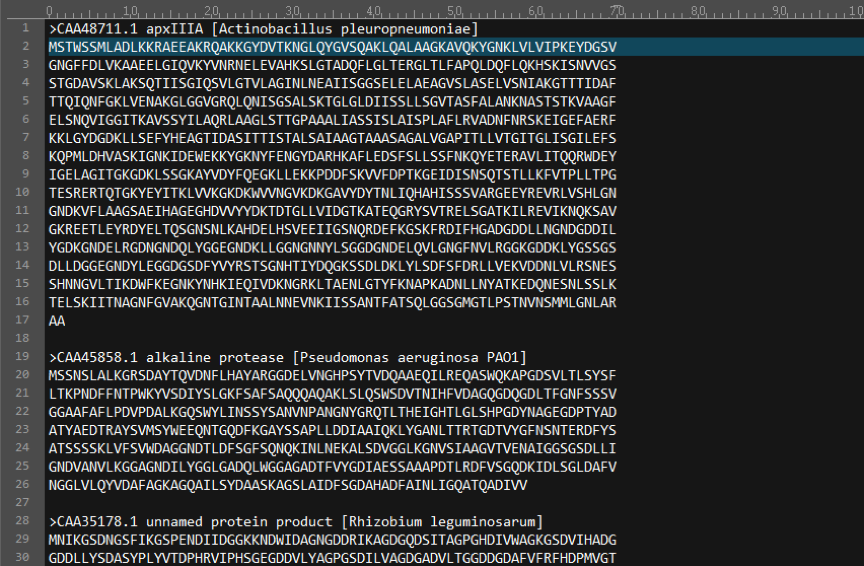
5) Running T1SEstacker in one command line:
get into ~/T1SEstacker/ through command line:
$ cd ~/T1SEstacker/
running T1SEstacker through command line:
$ perl T1SEstacker.pl test.fasta
6) Output file format:
The prediction results are given in ~/T1SEstacker. ‘T1SEstacker.out.txt’ gives the prediction results.
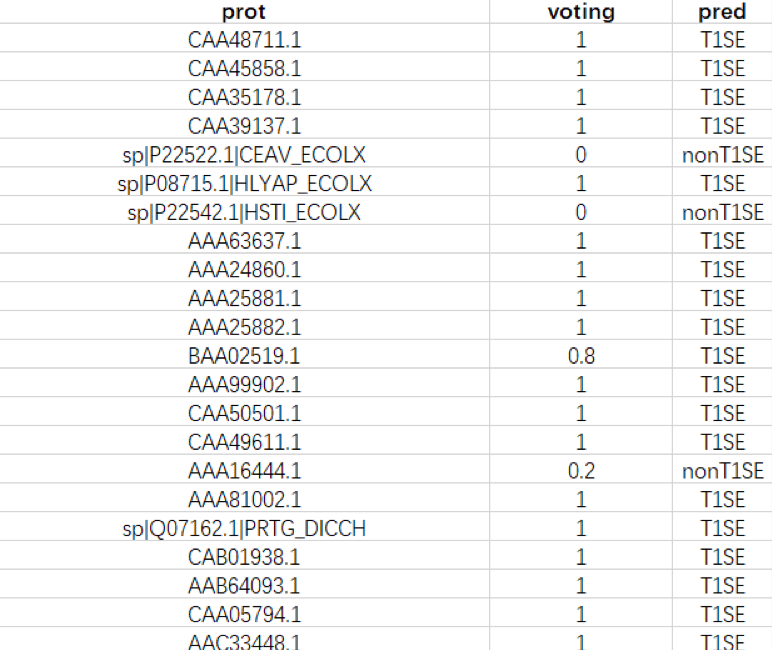
‘T1SEstacker.out.txt’ contains 3 columns: protein ID, the total score of five models, and the classification with default cutoff of each model and the classification according to the customized or default (0.6) cutoff.
Download User Guide