An Integrated Prediction Pipeline for Bacterial Type III Secreted Effectors.

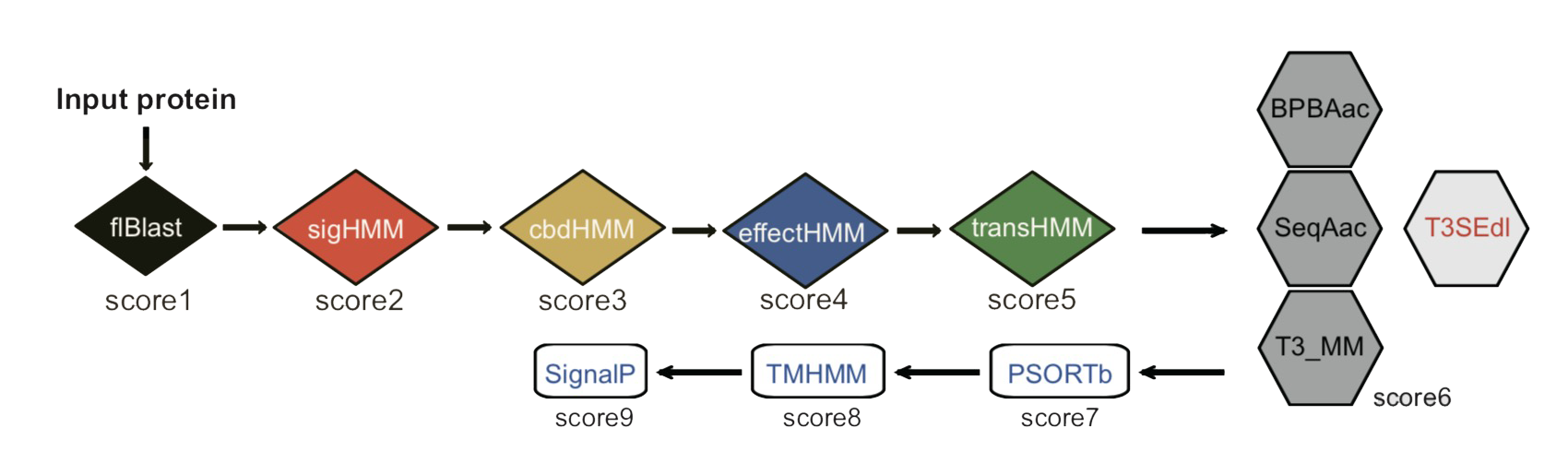
How to use the T3SEpp website to predict T3SS effecter proteins?
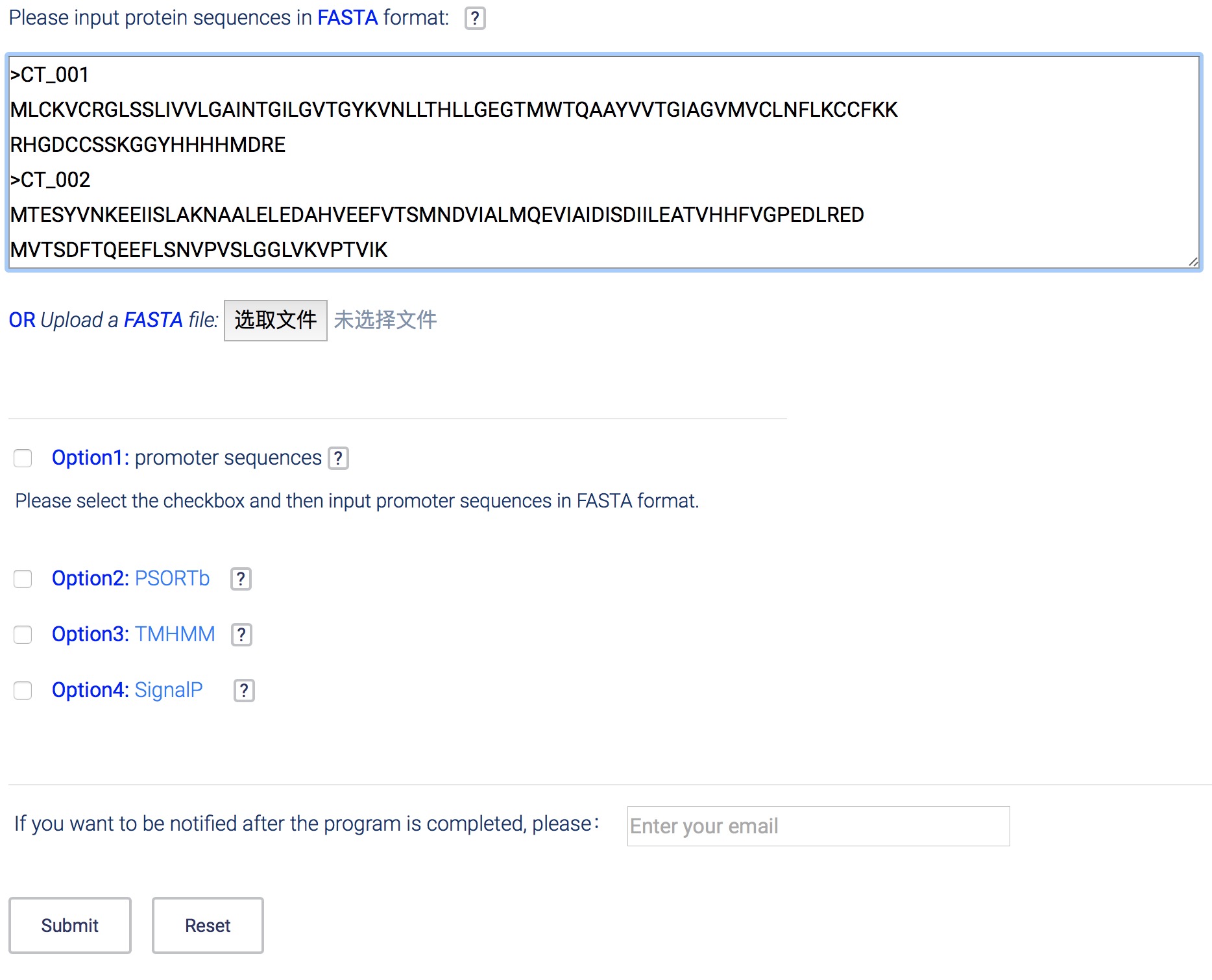
- Paste the candidate protein sequences into the textarea or upload a FASTA file (.fa/.fasta/.FASTA). For each protein, the name should neither be separated by space nor be with illegal characters such as "|".
- The T3SEpp website also offers 4 optional checkboxes, you can select them to upload other files to improve the prediction accuracy. Option2~4 need result-documents predicted by PSORTb, TMHMM and SignalP4.0, respectively. Details are described below.
- Offer an email address when you submit your job if you want to be informed once your job is finished.
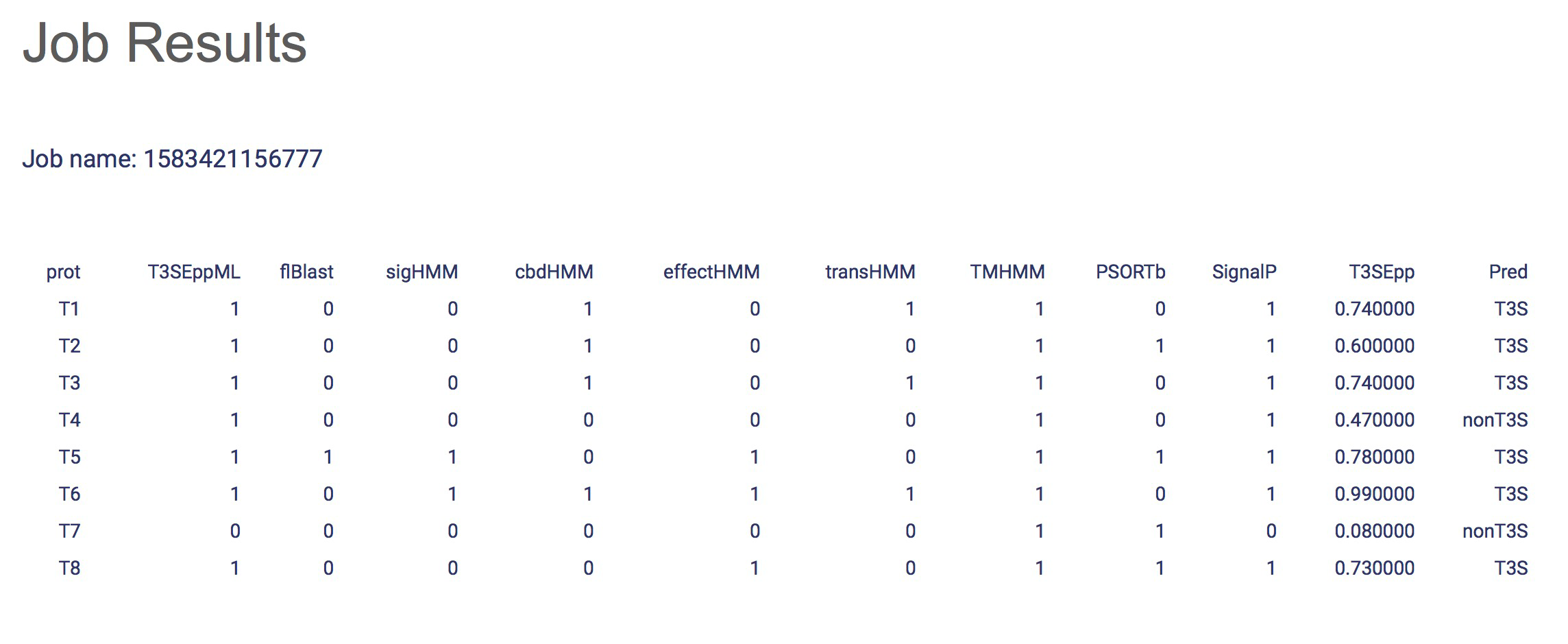
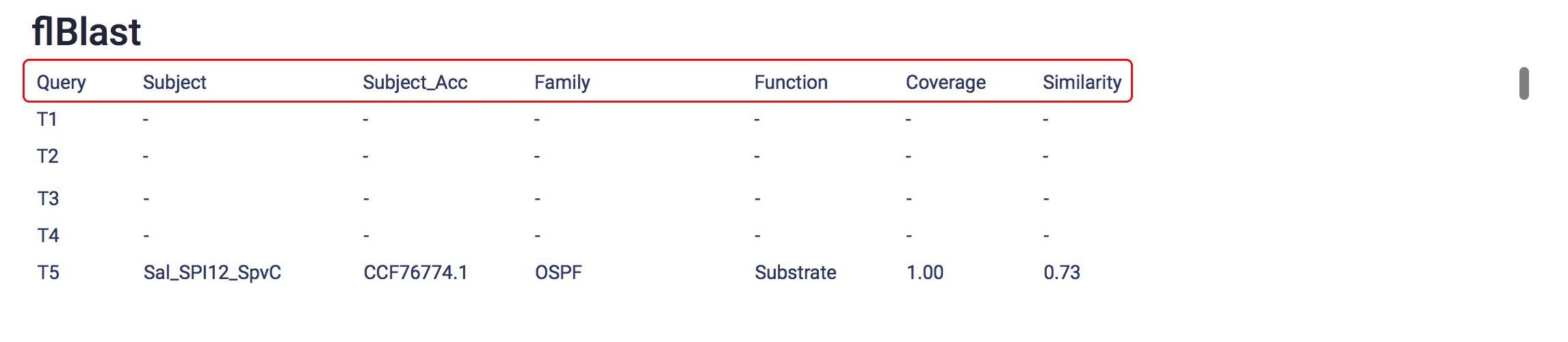
- An unique job name and a link will be provided after you submit your job successfully. You can check your job results by using the link.
- Standalone Version of T3SEpp is freely available at http://www.szu-bioinf.org/T3SEpp/download.html; and a manual of it can be download here.
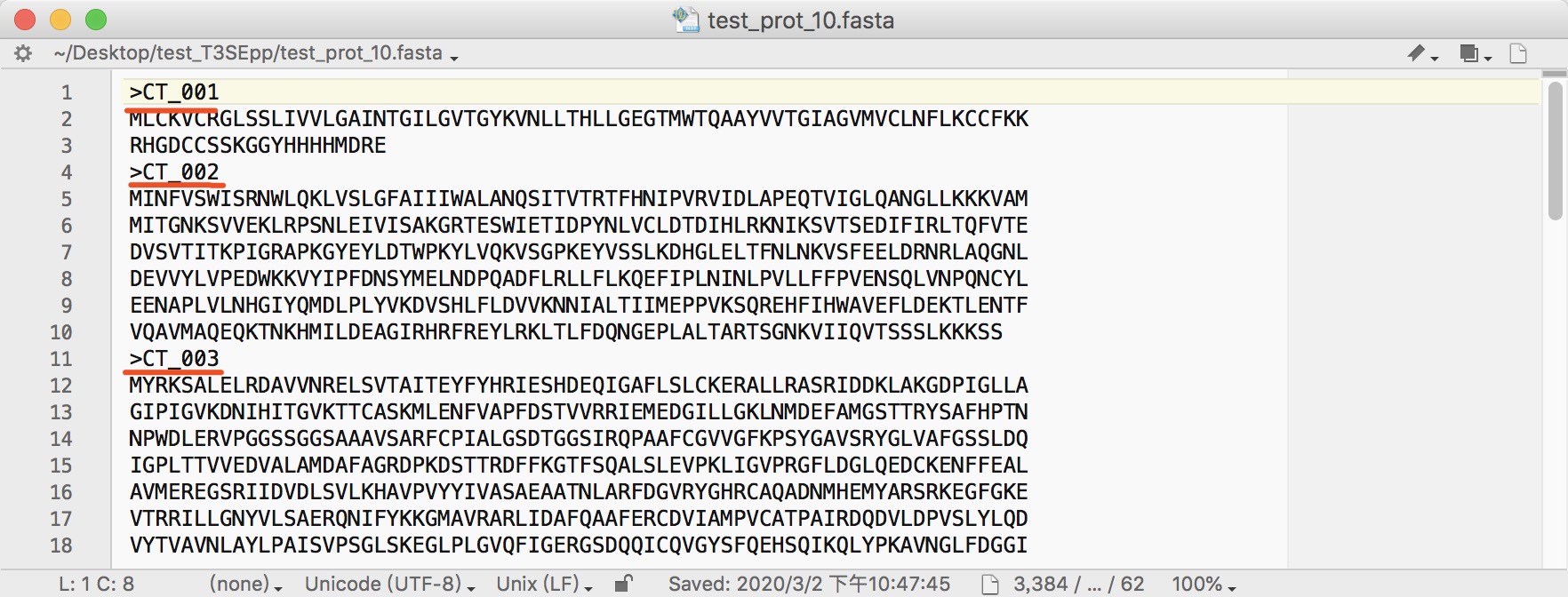

NOTE:
- Promoter sequences should be in FASTA format with a < 2001-nt length for each promoter.
- The name should exactly be consistent with that of protein sequence.
- The name should neither be separated by space nor be with illegal characters such as "|".
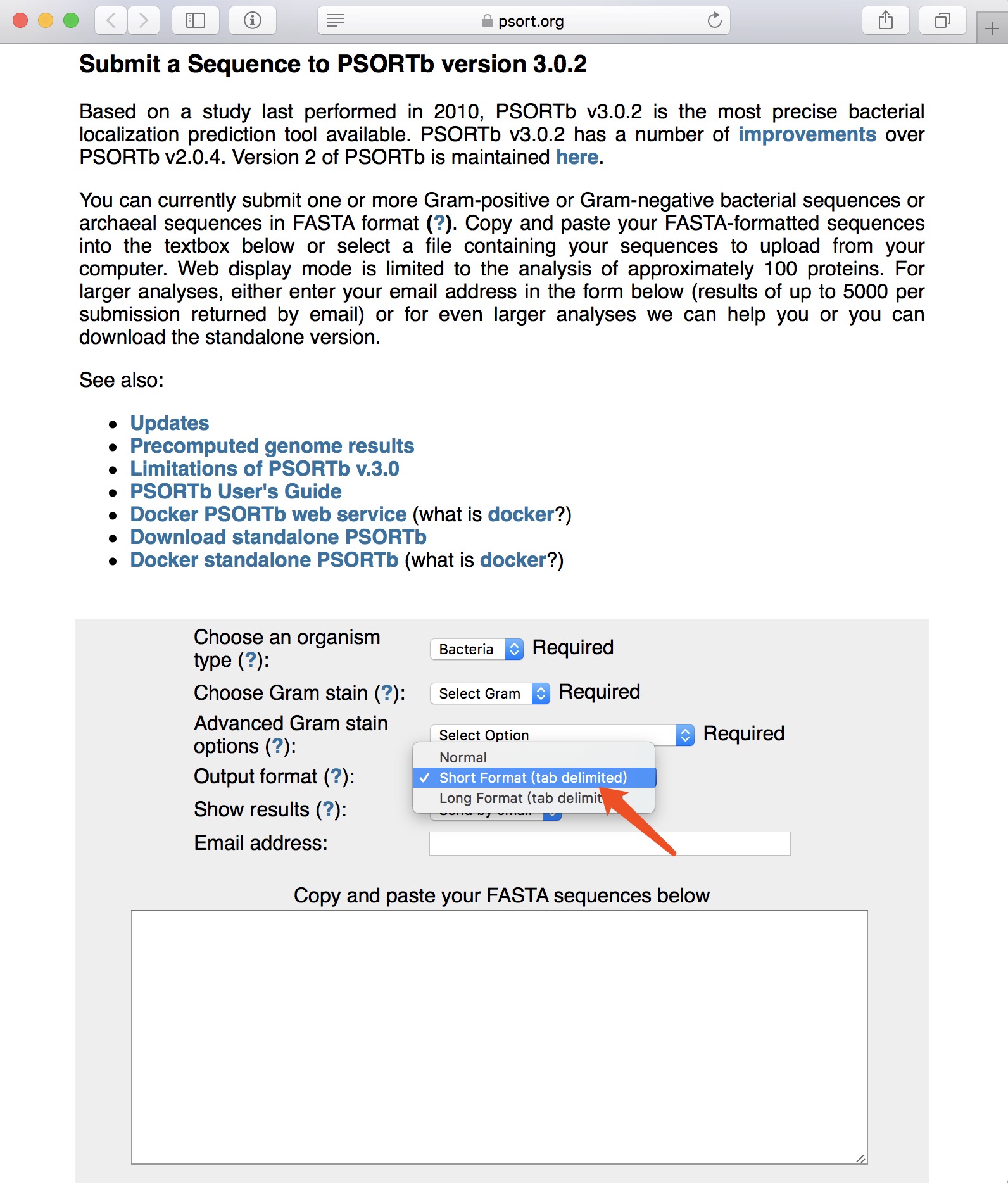
NOTE:
- PSORTb is available at https://www.psort.org/psortb/index.html.
- Using the Tab-delimited Output Format (Short Format) with 3 columns.
- Note that the protein name should exactly be consistent with that of protein sequence; the name should neither be separated by space nor be with illegal characters such as "|".
- Upload the TXT FILE without header.


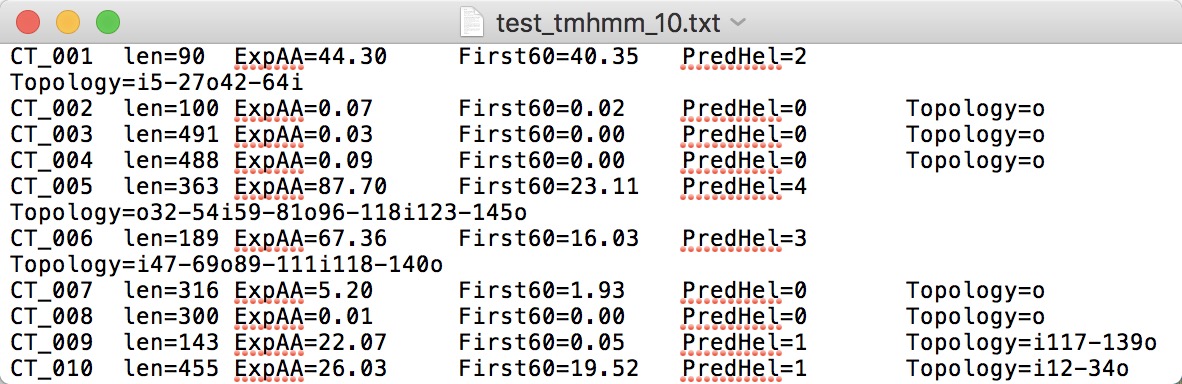
NOTE:
- TMHMM is available at https://services.healthtech.dtu.dk/service.php?TMHMM-2.0.
- Using the "One line per protein" output format.
- Note that the protein name should exactly be consistent with that of protein sequence; the name should neither be separated by space nor be with illegal characters such as "|".
- Upload the TXT FILE without header.
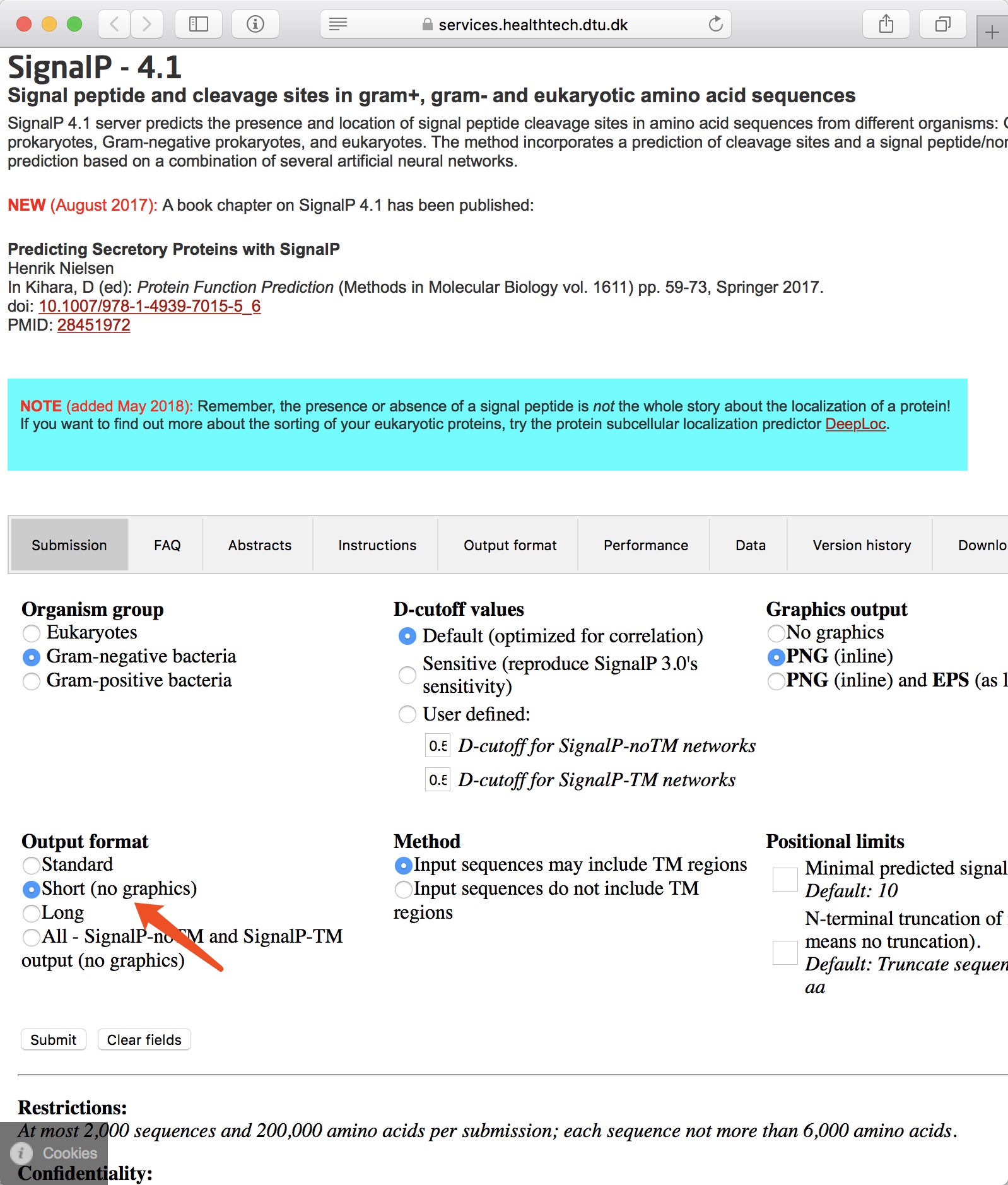
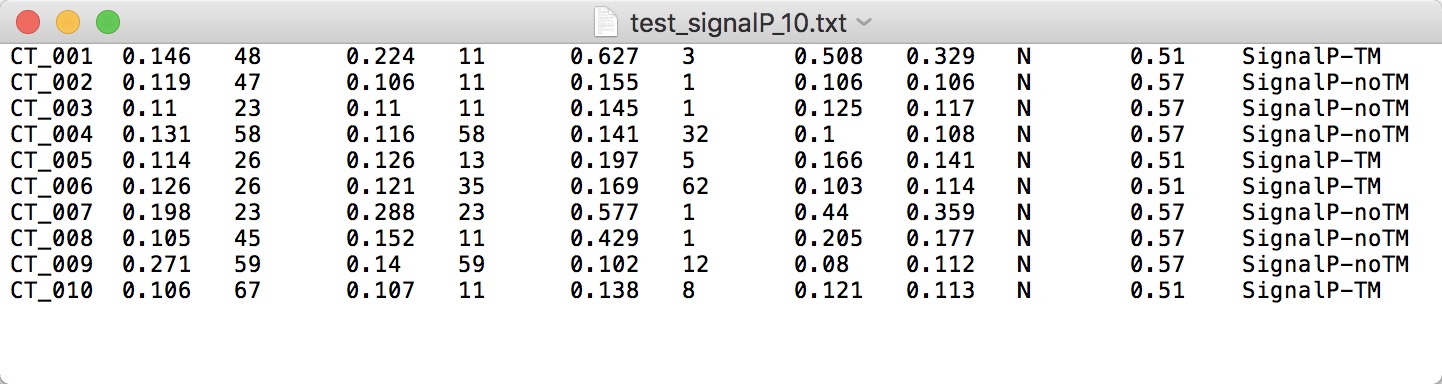
NOTE:
- SignalP is available at https://services.healthtech.dtu.dk/service.php?SignalP-4.1.
- Using the "Short (no graphics)" output format.
- Note that the protein name should exactly be consistent with that of protein sequence; the name should neither be separated by space nor be with illegal characters such as "|".
- Upload the TXT FILE without header.